The ratio of upper limit to lower limit is just the chi-square 0.975 quantile divided by the 0.025 quantile, it does not depend on the sample variance; checking this ratio for several degrees of freedom gives 27 degrees of freedom as the answer, which means that 28 observations are needed.
> data.frame(df=20:30,ratio=qchisq(.975,20:30)/qchisq(.025,20:30)) df ratio 1 20 3.562757 2 21 3.450280 3 22 3.349084 4 23 3.257514 5 24 3.174228 6 25 3.098120 7 26 3.028276 8 27 2.963932 9 28 2.904442 10 29 2.849260 11 30 2.797920
(a) Note that I gave up trying to match the tic-marks exactly.
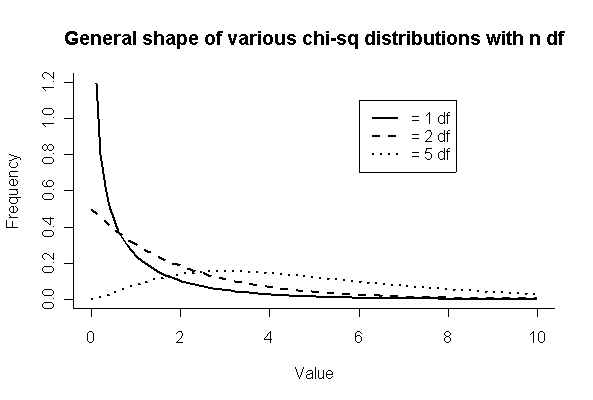
> xgr <- seq(0,10,len=100)
> plot(xgr,dchisq(xgr,1),type="l",xlim=c(0,10),ylim=c(0,1.2),
xlab="Value",ylab="Frequency",lwd=2,bty="l")
Warning message:
NaNs produced in: dchisq(x, df, log)
> lines(xgr,dchisq(xgr,2),lwd=2,lty=2)
> lines(xgr,dchisq(xgr,5),lwd=2,lty=3)
> legend(6,1.1,c("= 1 df","= 2 df","= 5 df"),lwd=2,lty=1:3)
> title("General shape of various chi-sq distributions with n df")

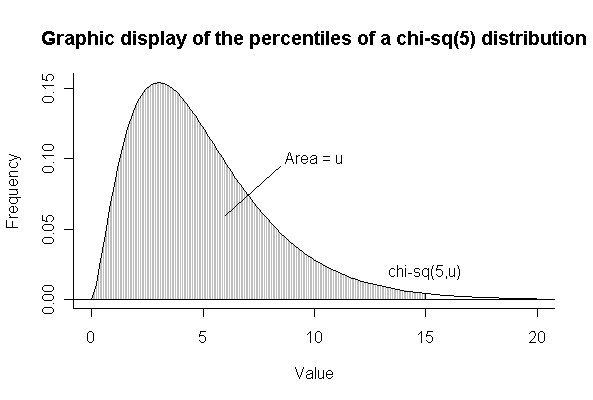
(b) Note that u = 0.9896377 from the pchisq(15,5) calculation below.
> xgr <- seq(0,20,len=100)
> xgr15 <- seq(0,15,len=200)
> plot(xgr,dchisq(xgr,5),xlab="Value",ylab="Frequency",type="l",bty="l")
> abline(h=0)
> lines(xgr15,dchisq(xgr15,5),type="h",col="gray")
> lines(15,dchisq(15,5),type="h")
> lines(xgr,dchisq(xgr,5))
> text(10,.1,"Area = u")
> lines(c(6,8.5),c(.06,.095))
> text(15,0.02,"chi-sq(5,u)")
> title("Graphic display of the percentiles of a chi-sq(5) distribution")
> pchisq(15,5)
[1] 0.9896377
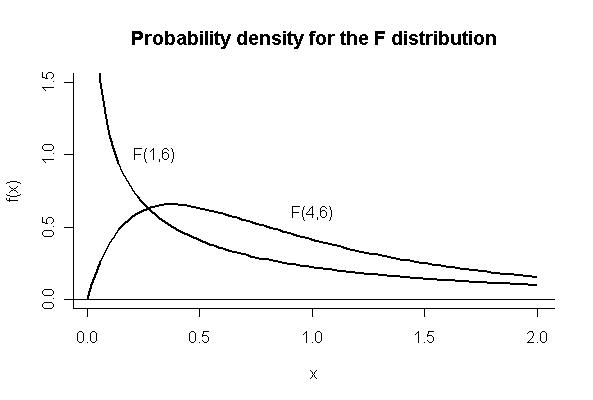
(c)
> xgr <- seq(0,2,len=100)
> plot(xgr[-1],df(xgr[-1],1,6),type="l",lwd=2,ylim=c(0,1.5),xlab="x",ylab="f(x)",bty="l")
> abline(h=0)
> lines(xgr,df(xgr,4,6),lwd=2)
> text(0.3,1,"F(1,6)")
> text(1,0.6,"F(4,6)")
> title("Probability density for the F distribution")
> 1975*310/(1627*255) [1] 1.47571
The odds for a boy are 47.6% higher for nonsmokers, compared to smokers.
> (1975/(1975+1627))/(255/(255+310)) [1] 1.214875
The probability of having a boy is 21.5% higher for nonsmokers, compared to smokers.
> (310/(255+310))/(1627/(1975+1627)) [1] 1.214701
The probability that both parents smoke is 21.5% higher for parents of a girl, compared to parents of a boy.
> sexrat <- chisq.test(matrix(c(1975, 1627, 255, 310), ncol = 2),correct=F)
> sexrat
Pearson's Chi-squared test
data: matrix(c(1975, 1627, 255, 310), ncol = 2)
X-squared = 18.4645, df = 1, p-value = 1.731e-05
> sexrat$obs
[,1] [,2]
[1,] 1975 255
[2,] 1627 310
> sexrat$exp
[,1] [,2]
[1,] 1927.636 302.3638
[2,] 1674.364 262.6362
> bmi1 base twoyr 1 26.5 26.1 2 26.1 26.4 3 25.4 25.5 4 27.4 26.3 5 25.4 NA 6 25.4 25.7 7 25.8 26.1 8 26.3 26.2 9 26.5 26.5 10 26.1 26.4 11 26.4 26.2 12 25.9 26.4 13 25.5 25.6 14 25.7 24.9 15 25.4 25.9 16 27.0 26.5
> t.test(bmi1$base,bmi1$twoyr,pair=T)
Paired t-test
data: bmi1$base and bmi1$twoyr
t = 0.3806, df = 14, p-value = 0.7092
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-0.2163206 0.3096539
sample estimates:
mean of the differences
0.04666667
> bmi1$base-bmi1$twoyr [1] 0.4 -0.3 -0.1 1.1 NA -0.3 -0.3 0.1 0.0 -0.3 0.2 -0.5 -0.1 0.8 [15] -0.5 0.5 > 2*pbinom(6,14,.5) [1] 0.7905273
> anova(lm(twoyr~base,data=bmi1))
Analysis of Variance Table
Response: twoyr
Df Sum Sq Mean Sq F value Pr(>F)
base 1 1.1218 1.1218 8.4025 0.01244 *
Residuals 13 1.7356 0.1335
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
> summary(lm(twoyr~base,data=bmi1))
Call:
lm(formula = twoyr ~ base, data = bmi1)
Residuals:
Min 1Q Median 3Q Max
-0.96164 -0.15276 0.02683 0.22668 0.44428
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 13.7721 4.2356 3.252 0.00631 **
base 0.4704 0.1623 2.899 0.01244 *
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
Residual standard error: 0.3654 on 13 degrees of freedom
Multiple R-Squared: 0.3926, Adjusted R-squared: 0.3459
F-statistic: 8.402 on 1 and 13 DF, p-value: 0.01244
> plot(rep(0,16),bmi1$base-bmi1$twoyr)
> boxplot(bmi1$base-bmi1$twoyr)
> title(ylab="base - twoyr")
> hist(bmi1$base-bmi1$twoyr)
> plot(bmi1$base,bmi1$twoyr)
> abline(0,1)
> abline(lm(twoyr~base,data=bmi1), lty=2)
|
|
|
|
|
|
> t.test(creek$pH[creek$loc=="Up"],creek$pH[creek$loc=="Down"],var=T)
Two Sample t-test
data: creek$pH[creek$loc == "Up"] and creek$pH[creek$loc == "Down"]
t = -1.133, df = 18, p-value = 0.2721
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-1.7325194 0.5185194
sample estimates:
mean of x mean of y
6.461 7.068
> anova(lm(pH~location,data=creek))
Analysis of Variance Table
Response: pH
Df Sum Sq Mean Sq F value Pr(>F)
location 1 1.8422 1.8422 1.2838 0.2721
Residuals 18 25.8303 1.4350
> F0 <- var(creek$pH[creek$loc=="Up"])/var(creek$pH[creek$loc=="Down"])
> F0
[1] 8.43619
> 2*(1-pf(F0,9,9))
[1] 0.003964524
> var(creek$pH[creek$loc=="Up"])
[1] 2.565877
> var(creek$pH[creek$loc=="Down"])
[1] 0.3041511
> boxplot(split(creek$pH,creek$location))
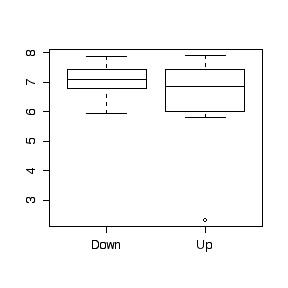
> var(creek$pH[creek$loc=="Up"][-3])
[1] 0.5049611
> F0a <- var(creek$pH[creek$loc=="Up"][-3])/var(creek$pH[creek$loc=="Down"])
> F0a
[1] 1.660231
> 2*(1-pf(F0a,8,9))
[1] 0.4654831
> anova(lm(pH~location,data=creek[-3,]))
Analysis of Variance Table
Response: pH
Df Sum Sq Mean Sq F value Pr(>F)
location 1 0.1022 0.1022 0.2564 0.6191
Residuals 17 6.7770 0.3986
All of the required answers are found in the output below. There is, of course, no evidence (P = 0.56 by the F- test in the ANOVA or by the t-test on the slope) of a relationship between % reticulytes and lymphocytes per mm2. This is confirmed by the R2 which is near 0, which means that linear prediction is not possible.
The mean square residual gives s2y.x = 490818.
> anemia
retic lymph
1 3.6 1700
2 2.0 3078
3 0.3 1820
4 0.3 2706
5 0.2 2086
6 3.0 2299
7 0.0 676
8 1.0 2088
9 2.2 2013
> fitanemia<-lm(lymph~retic,anemia)
> plot(lymph~retic,anemia)
> abline(fitanemia)
> anova(fitanemia)
Analysis of Variance Table
Response: lymph
Df Sum Sq Mean Sq F value Pr(>F)
retic 1 180750 180750 0.3683 0.5631
Residuals 7 3435727 490818
> summary(fitanemia)
Call:
lm(formula = lymph ~ retic, data = anemia)
Residuals:
Min 1Q Median 3Q Max
-1218.82 -128.47 67.84 168.76 958.95
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1894.8 348.5 5.437 0.000969 ***
retic 112.1 184.7 0.607 0.563109
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
Residual standard error: 700.6 on 7 degrees of freedom
Multiple R-Squared: 0.04998, Adjusted R-squared: -0.08574
F-statistic: 0.3683 on 1 and 7 DF, p-value: 0.5631
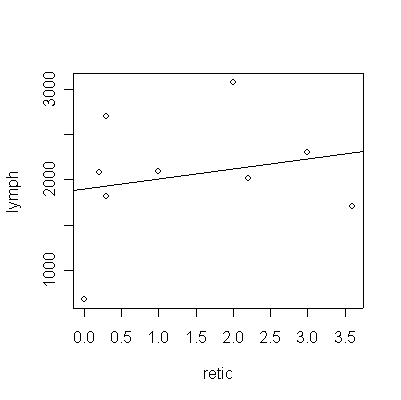
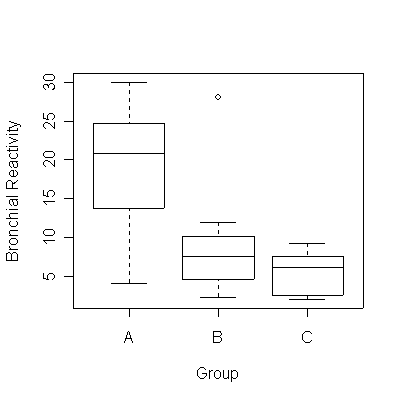
There is some evidence (P = 0.018) that the mean bronchial reactivity varies between the three groups based on the ratio FEV1/FVC. Note that the graph suggests that Group A has greater variance than B or C but the sample sizes are too small to be sure.
> lung
react group
1 20.8 A
2 4.1 A
3 30.0 A
4 24.7 A
5 13.8 A
6 7.5 B
7 7.5 B
8 11.9 B
9 4.5 B
10 3.1 B
11 8.0 B
12 4.7 B
13 28.1 B
14 10.3 B
15 10.0 B
16 5.1 B
17 2.2 B
18 9.2 C
19 2.0 C
20 2.5 C
21 6.1 C
22 7.5 C
> boxplot(react~group,lung,xlab="Group",ylab="Bronchial Reactivity")
> anova(lm(react~group,lung))
Analysis of Variance Table
Response: react
Df Sum Sq Mean Sq F value Pr(>F)
group 2 503.55 251.77 4.9893 0.01813 *
Residuals 19 958.80 50.46
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
> toxic1
life poison antidote
1 0.31 I A
2 0.46 I A
3 0.36 II A
4 0.40 II A
5 0.22 III A
6 0.18 III A
7 0.82 I B
8 0.88 I B
9 0.92 II B
10 0.49 II B
11 0.30 III B
12 0.38 III B
13 0.43 I C
14 0.63 I C
15 0.44 II C
16 0.31 II C
17 0.23 III C
18 0.24 III C
19 0.45 I D
20 0.66 I D
21 0.56 II D
22 0.71 II D
23 0.30 III D
24 0.31 III D
> anova(lm(life~poison*antidote, data=toxic1))
Analysis of Variance Table
Response: life
Df Sum Sq Mean Sq F value Pr(>F)
poison 2 0.43641 0.21820 15.2103 0.0005124 ***
antidote 3 0.33875 0.11292 7.8709 0.0036184 **
poison:antidote 6 0.08989 0.01498 1.0443 0.4445361
Residuals 12 0.17215 0.01435
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
> mse <- anova(lm(life~poison*antidote, data=toxic1))$"Mean Sq"[4]
> mse
0.01434583
> mse/c(qchisq(.995,12)/12, qchisq(.005,12)/12)
[1] 0.006083142 0.056005165
|
|
|
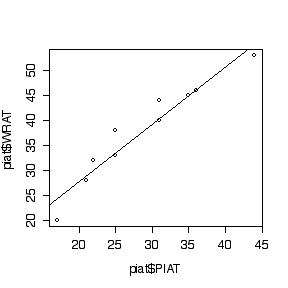
> plot(piat$PIAT,piat$WRAT)
> abline(lm(WRAT~PIAT,data=piat))
> summary(lm(WRAT~PIAT,data=piat))
Call:
lm(formula = WRAT ~ PIAT, data = piat)
Residuals:
Min 1Q Median 3Q Max
-4.4511 -0.9485 -0.4175 1.3157 4.3531
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4.9099 3.3642 1.459 0.183
PIAT 1.1495 0.1131 10.161 7.53e-06 ***
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
Residual standard error: 2.785 on 8 degrees of freedom
Multiple R-Squared: 0.9281, Adjusted R-squared: 0.9191
F-statistic: 103.2 on 1 and 8 DF, p-value: 7.535e-06
> anova(lm(WRAT~PIAT,data=piat))
Analysis of Variance Table
Response: WRAT
Df Sum Sq Mean Sq F value Pr(>F)
PIAT 1 800.84 800.84 103.24 7.535e-06 ***
Residuals 8 62.06 7.76
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
> anova(lm(WRAT~PIAT+as.factor(PIAT),data=piat))
Analysis of Variance Table
Response: WRAT
Df Sum Sq Mean Sq F value Pr(>F)
PIAT 1 800.84 800.84 78.1310 0.01256 *
as.factor(PIAT) 6 41.56 6.93 0.6757 0.69970
Residuals 2 20.50 10.25
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
> mse1 <- anova(lm(WRAT~PIAT,data=piat))$"Mean Sq"[2]
> mse2 <- anova(lm(WRAT~PIAT+as.factor(PIAT),data=piat))$"Mean Sq"[3]
> mse1
7.757136
> mse1/c(qchisq(.995,8)/8,qchisq(.005,8)/8)
[1] 2.826564 46.159240
> mse2
10.25
> mse2/c(qchisq(.995,2)/2,qchisq(.005,2)/2)
[1] 1.934576 2044.870718
> predict(lm(WRAT~PIAT,data=piat),data.frame(PIAT=30))
[1] 39.39432