[Graph 3]
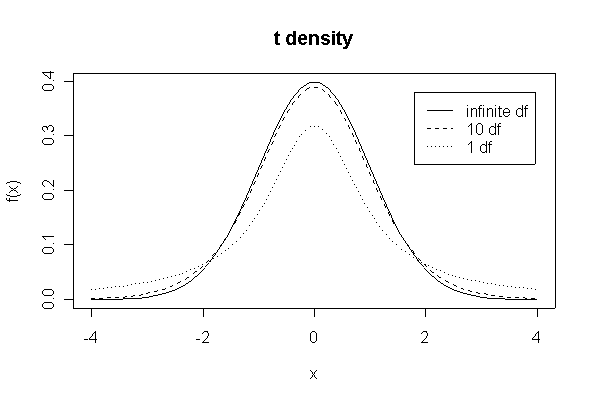
> xgr <- seq(-4,4,length=50)
> plot(xgr, dnorm(xgr), type = "l", lty = 1, xlab = "x", ylab ="f(x)")
> lines(xgr,dt(xgr,10),lty=2)
> lines(xgr,dt(xgr,1),lty=3)
> legend(1.8,.38,c("infinite df","10 df","1 df"),lty=1:3)
> title("t density")

[Graph 3]
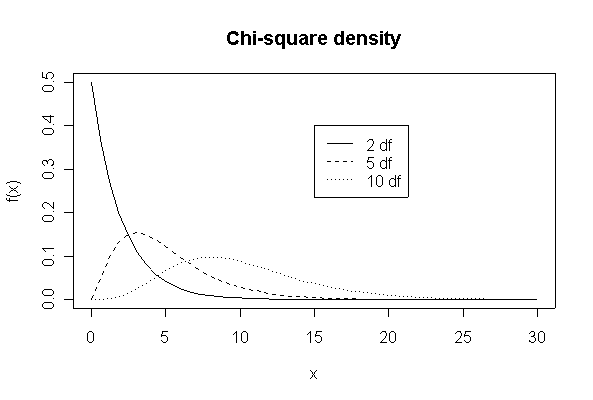
> xgr <- seq(0,30,length=50)
> plot(xgr, dchisq(xgr, 2), type = "l", lty = 1, xlab = "x", ylab ="f(x)")
> lines(xgr,dchisq(xgr,5),lty=2)
> lines(xgr,dchisq(xgr,10),lty=3)
> legend(15,.4,c("2 df","5 df","10 df"),lty=1:3)
> title("Chi-square density")
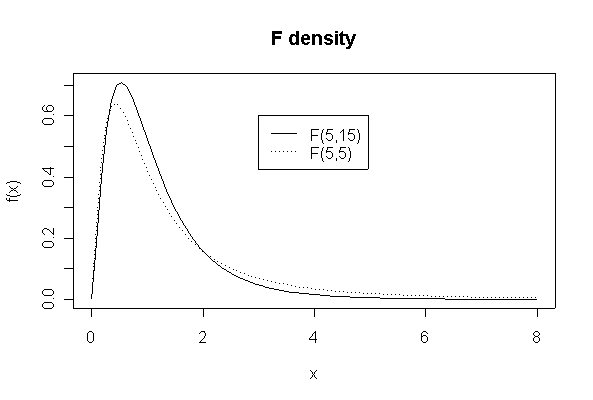
[Graph 3]
> xgr <- seq(0,8,length=90)
> plot(xgr, df(xgr,5,15), type = "l", lty = 1, xlab = "x", ylab ="f(x)")
> lines(xgr,df(xgr,5,5),lty=3)
> legend(3,.6,c("F(5,15)","F(5,5)"),lty=c(1,3))
> title("F density")
[8 marks]
When n = 5 the coverage is only about 86%, much less than the 95% intended. When n = 100 the coverage is 95%. If n > 40 it appears that the coverage will be close enough to 95%.
> n <- 5
> expdata <- matrix(rexp(1000*n, 1/10), ncol=n)
> xbare <- apply(expdata, 1, mean)
> sxe <- sqrt(apply(expdata, 1, var))
> llim <- xbare-qt(.975,n-1)*sxe/sqrt(n)
> ulim <- xbare+qt(.975,n-1)*sxe/sqrt(n)
> mean(10 > llim & 10 < ulim)
[1] 0.864
> plot(c(0,20),c(-10,40),type="n",xlab="Interval Number",ylab="x")
> for (i in 1:20)
{ points(i, xbare[i], pch=16)
lines(c(i,i), c(llim[i],ulim[i])) }
> abline(h=10, lty=3)
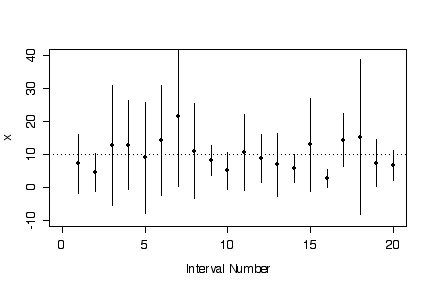 > n <- 10
> expdata <- matrix(rexp(1000*n, 1/10), ncol=n)
> xbare <- apply(expdata, 1, mean)
> sxe <- sqrt(apply(expdata, 1, var))
> llim <- xbare-qt(.975,n-1)*sxe/sqrt(n)
> ulim <- xbare+qt(.975,n-1)*sxe/sqrt(n)
> mean(10 > llim & 10 < ulim)
[1] 0.92
> n <- 20
> expdata <- matrix(rexp(1000*n, 1/10), ncol=n)
> xbare <- apply(expdata, 1, mean)
> sxe <- sqrt(apply(expdata, 1, var))
> llim <- xbare-qt(.975,n-1)*sxe/sqrt(n)
> ulim <- xbare+qt(.975,n-1)*sxe/sqrt(n)
> mean(10 > llim & 10 < ulim)
[1] 0.924
> n <- 40
> expdata <- matrix(rexp(1000*n, 1/10), ncol=n)
> xbare <- apply(expdata, 1, mean)
> sxe <- sqrt(apply(expdata, 1, var))
> llim <- xbare-qt(.975,n-1)*sxe/sqrt(n)
> ulim <- xbare+qt(.975,n-1)*sxe/sqrt(n)
> mean(10 > llim & 10 < ulim)
[1] 0.935
> n <- 100
> expdata <- matrix(rexp(1000*n, 1/10), ncol=n)
> xbare <- apply(expdata, 1, mean)
> sxe <- sqrt(apply(expdata, 1, var))
> llim <- xbare-qt(.975,n-1)*sxe/sqrt(n)
> ulim <- xbare+qt(.975,n-1)*sxe/sqrt(n)
> mean(10 > llim & 10 < ulim)
[1] 0.952
> n <- 10
> expdata <- matrix(rexp(1000*n, 1/10), ncol=n)
> xbare <- apply(expdata, 1, mean)
> sxe <- sqrt(apply(expdata, 1, var))
> llim <- xbare-qt(.975,n-1)*sxe/sqrt(n)
> ulim <- xbare+qt(.975,n-1)*sxe/sqrt(n)
> mean(10 > llim & 10 < ulim)
[1] 0.92
> n <- 20
> expdata <- matrix(rexp(1000*n, 1/10), ncol=n)
> xbare <- apply(expdata, 1, mean)
> sxe <- sqrt(apply(expdata, 1, var))
> llim <- xbare-qt(.975,n-1)*sxe/sqrt(n)
> ulim <- xbare+qt(.975,n-1)*sxe/sqrt(n)
> mean(10 > llim & 10 < ulim)
[1] 0.924
> n <- 40
> expdata <- matrix(rexp(1000*n, 1/10), ncol=n)
> xbare <- apply(expdata, 1, mean)
> sxe <- sqrt(apply(expdata, 1, var))
> llim <- xbare-qt(.975,n-1)*sxe/sqrt(n)
> ulim <- xbare+qt(.975,n-1)*sxe/sqrt(n)
> mean(10 > llim & 10 < ulim)
[1] 0.935
> n <- 100
> expdata <- matrix(rexp(1000*n, 1/10), ncol=n)
> xbare <- apply(expdata, 1, mean)
> sxe <- sqrt(apply(expdata, 1, var))
> llim <- xbare-qt(.975,n-1)*sxe/sqrt(n)
> ulim <- xbare+qt(.975,n-1)*sxe/sqrt(n)
> mean(10 > llim & 10 < ulim)
[1] 0.952
[8 marks]
The sample mean and variance from normal data are statistically independent, as demonstrated by the scatterplot below. This means that t confidence intervals from normal data will be too wide or too narrow independently of whether they are to low or too high. Exponential data, however, give positively correlated sample mean and variance. This is because, being positively skewed, if there happen to be observations from the right tail in a sample, the sample mean and the sample variance will both be inflated. The result is that the t-based confidence intervals that are too high will tend to be too wide (and hence more likely to include the true value) and those that are too low will tend to be too narrow (and hence more likely to miss the true value).
|
|
|
(a) [5 marks]
> qchisq(.975,27)/qchisq(.025,27) [1] 2.963932 > qchisq(.975,26)/qchisq(.025,26) [1] 3.028276
Hence 27 degrees of freedom are required.
(b) [10 marks]
The number of pathogens found in a sample will ,with probability 0.7, be Poisson with mean 30, and, with probability 0.3, be Poisson with mean 60. A Bayes Theorem calculation gives the conditional probability of being at the higher mean, given that 40 pathogens were observed.
> .3*dpois(40,60)/(.3*dpois(40,60)+.7*dpois(40,30)) [1] 0.04223268
(a) [valid analysis 2, graph 3, analysis 4, assumptions 2, conclusions 1]
> hex
indoor outdoor
1 0.22 0.90
2 0.18 0.66
3 0.28 1.24
4 0.34 0.37
5 0.18 1.55
6 0.12 0.54
7 0.29 0.27
8 0.08 0.68
9 0.39 1.26
10 0.28 0.48
> anova(lm(indoor~outdoor,hex))
Analysis of Variance Table
Response: indoor
Df Sum Sq Mean Sq F value Pr(>F)
outdoor 1 0.000153 0.000153 0.0144 0.9076
Residuals 8 0.085487 0.010686
> summary(lm(indoor~outdoor,hex))
Call:
lm(formula = indoor ~ outdoor, data = hex)
Residuals:
Min 1Q Median 3Q Max
-0.15489 -0.06114 0.01135 0.05606 0.14951
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.228326 0.071893 3.176 0.0131 *
outdoor 0.009652 0.080542 0.120 0.9076
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
Residual standard error: 0.1034 on 8 degrees of freedom
Multiple R-Squared: 0.001792, Adjusted R-squared: -0.123
F-statistic: 0.01436 on 1 and 8 DF, p-value: 0.9076
> plot(indoor~outdoor,hex)
> abline(lm(indoor~outdoor,hex))
> t.test(hex$ind,hex$outd,pair=T) Paired t-test data: hex$ind and hex$outd t = -4.066, df = 9, p-value = 0.002816 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: -0.8700034 -0.2479966 sample estimates: mean of the differences -0.559 > stem(hex$ind-hex$outd) The decimal point is at the | -1 | 40 -0 | 9765 -0 | 420 0 | 0
(b) [valid analysis 2, graph 3, analysis 5, assumptions 2, conclusions 1]
> wafer
defects clean
1 53 S
2 193 S
3 113 S
4 640 S
5 800 S
6 140 S
7 85 S
8 658 S
9 140 S
10 140 S
11 26 M
12 90 M
13 546 M
14 90 M
15 120 M
> t.test(defects~clean,wafer,var.eq=T)
Two Sample t-test
data: defects by clean
t = -0.8445, df = 13, p-value = 0.4137
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-433.3820 189.7820
sample estimates:
mean in group M mean in group S
174.4 296.2
> boxplot(defects~clean,wafer)
> var(wafer$defects[wafer$clean=="S"])/var(wafer$defects[wafer$clean=="M"])
[1] 1.814901
> 2*pf(var(wafer$defects[wafer$clean=="S"])/var(wafer$defects[wafer$clean=="M"]),9,4,lower.tail=F)
[1] 0.5929594
[either interaction plot 5, ANOVA with interaction 14, CI for residual variance 3, conclusions 3]
> paper
pctwr etime temp
1 24 10 100
2 26 10 100
3 21 10 100
4 25 10 100
5 39 20 100
6 34 20 100
7 37 20 100
8 40 20 100
9 58 30 100
10 55 30 100
11 56 30 100
12 53 30 100
13 33 10 120
14 33 10 120
15 36 10 120
16 32 10 120
17 51 20 120
18 50 20 120
19 47 20 120
20 52 20 120
21 75 30 120
22 71 30 120
23 70 30 120
24 73 30 120
25 45 10 140
26 49 10 140
27 44 10 140
28 45 10 140
29 67 20 140
30 64 20 140
31 68 20 140
32 65 20 140
33 89 30 140
34 87 30 140
35 86 30 140
36 83 30 140
> anova(lm(pctwr~as.factor(etime)*as.factor(temp), paper))
Analysis of Variance Table
Response: pctwr
Df Sum Sq Mean Sq F value Pr(>F)
as.factor(etime) 2 8200.4 4100.2 856.5203 < 2.2e-16 ***
as.factor(temp) 2 4376.7 2188.4 457.1431 < 2.2e-16 ***
as.factor(etime):as.factor(temp) 4 103.3 25.8 5.3936 0.002522 **
Residuals 27 129.2 4.8
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
> interactplot(paper$pctwr,as.factor(paper$etime),as.factor(paper$temp))
> interactplot(paper$pctwr,as.factor(paper$temp),as.factor(paper$etime))
> anova(lm(pctwr~as.factor(etime)*as.factor(temp),paper))[4, 3]
4.787037
> anova(lm(pctwr~as.factor(etime)*as.factor(temp),paper))[4, 3]/(qchisq(c(.975,.025),27)/27)
[1] 2.992278 8.868909
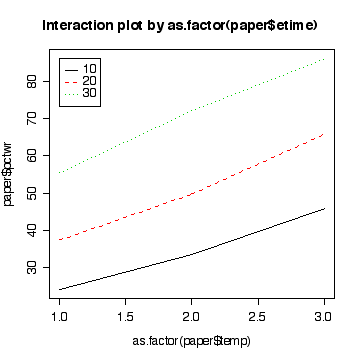
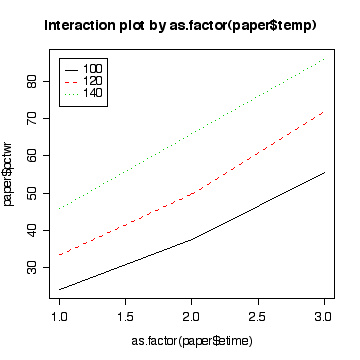
[ANOVA with lack of fit 13, CI for residual variance 3, assumptions 3, conclusions 3, predictions and comment 3]
> summary(lm(pctwr~etime, paper[paper$temp==100,]))
Call:
lm(formula = pctwr ~ etime, data = paper[paper$temp == 100, ])
Residuals:
Min 1Q Median 3Q Max
-5.000 -1.812 0.500 1.375 3.250
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 7.50000 1.89407 3.96 0.00269 **
etime 1.57500 0.08768 17.96 6.12e-09 ***
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
Residual standard error: 2.48 on 10 degrees of freedom
Multiple R-Squared: 0.9699, Adjusted R-squared: 0.9669
F-statistic: 322.7 on 1 and 10 DF, p-value: 6.116e-09
> anova(lm(pctwr~etime, paper[paper$temp==100,]))
Analysis of Variance Table
Response: pctwr
Df Sum Sq Mean Sq F value Pr(>F)
etime 1 1984.50 1984.50 322.68 6.116e-09 ***
Residuals 10 61.50 6.15
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
> anova(lm(pctwr~etime, paper[paper$temp==100,]))[2,3]
6.15
> anova(lm(pctwr~etime, paper[paper$temp==100,]))[2,3]/(qchisq(c(.975,.025),10)/10)
[1] 3.002464 18.940719
> anova(lm(pctwr~etime+as.factor(etime), paper[paper$temp==100,]))
Analysis of Variance Table
Response: pctwr
Df Sum Sq Mean Sq F value Pr(>F)
etime 1 1984.50 1984.50 372.0937 1.249e-08 ***
as.factor(etime) 1 13.50 13.50 2.5313 0.1461
Residuals 9 48.00 5.33
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
> anova(lm(pctwr~etime+as.factor(etime), paper[paper$temp==100,]))[3,3]
5.333333
> anova(lm(pctwr~etime+as.factor(etime), paper[paper$temp==100,]))[3,3]/(qchisq(c(.975,.025),9)/9)
[1] 2.523292 17.775214
> predict(lm(pctwr~etime, paper[paper$temp==100,]), newdata=data.frame(etime=c(0,25,60)))
1 2 3
7.500 46.875 102.000
> plot(pctwr~etime, paper[paper$temp==100,]) > abline(lm(pctwr~etime, paper[paper$temp==100,]))
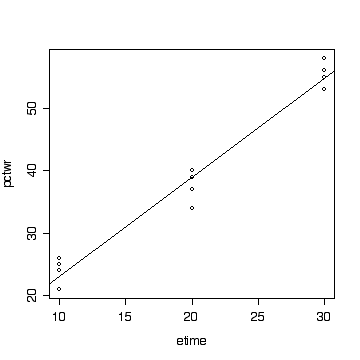
[ANOVA with interaction 12, conclusions 3, plots of residuals and discussion 10]
> baked
density pos temp posf tempf
1 570 1 800 1 800
2 565 1 800 1 800
3 583 1 800 1 800
4 1063 1 825 1 825
5 1080 1 825 1 825
6 1043 1 825 1 825
7 565 1 850 1 850
8 510 1 850 1 850
9 590 1 850 1 850
10 528 2 800 2 800
11 547 2 800 2 800
12 521 2 800 2 800
13 988 2 825 2 825
14 1026 2 825 2 825
15 1004 2 825 2 825
16 526 2 850 2 850
17 538 2 850 2 850
18 532 2 850 2 850
> anova(lm(density~posf*tempf, baked))
Analysis of Variance Table
Response: density
Df Sum Sq Mean Sq F value Pr(>F)
posf 1 7160 7160 15.998 0.001762 **
tempf 2 945342 472671 1056.117 3.25e-14 ***
posf:tempf 2 818 409 0.914 0.427110
Residuals 12 5371 448
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
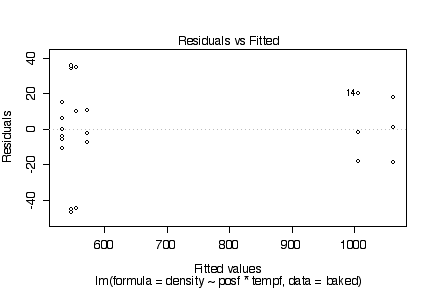
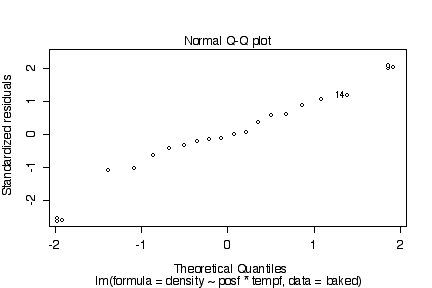
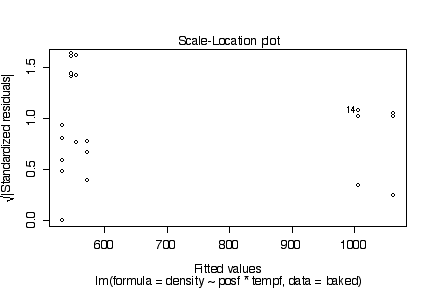
There are no obvious systematic patterns in the residuals. This suggests that the model is adequate and the homoscedasticity assumption is reasonable. The probability plot is straight enough to validate the assumpton of normality.
> plot(lm(density~posf*tempf, baked))